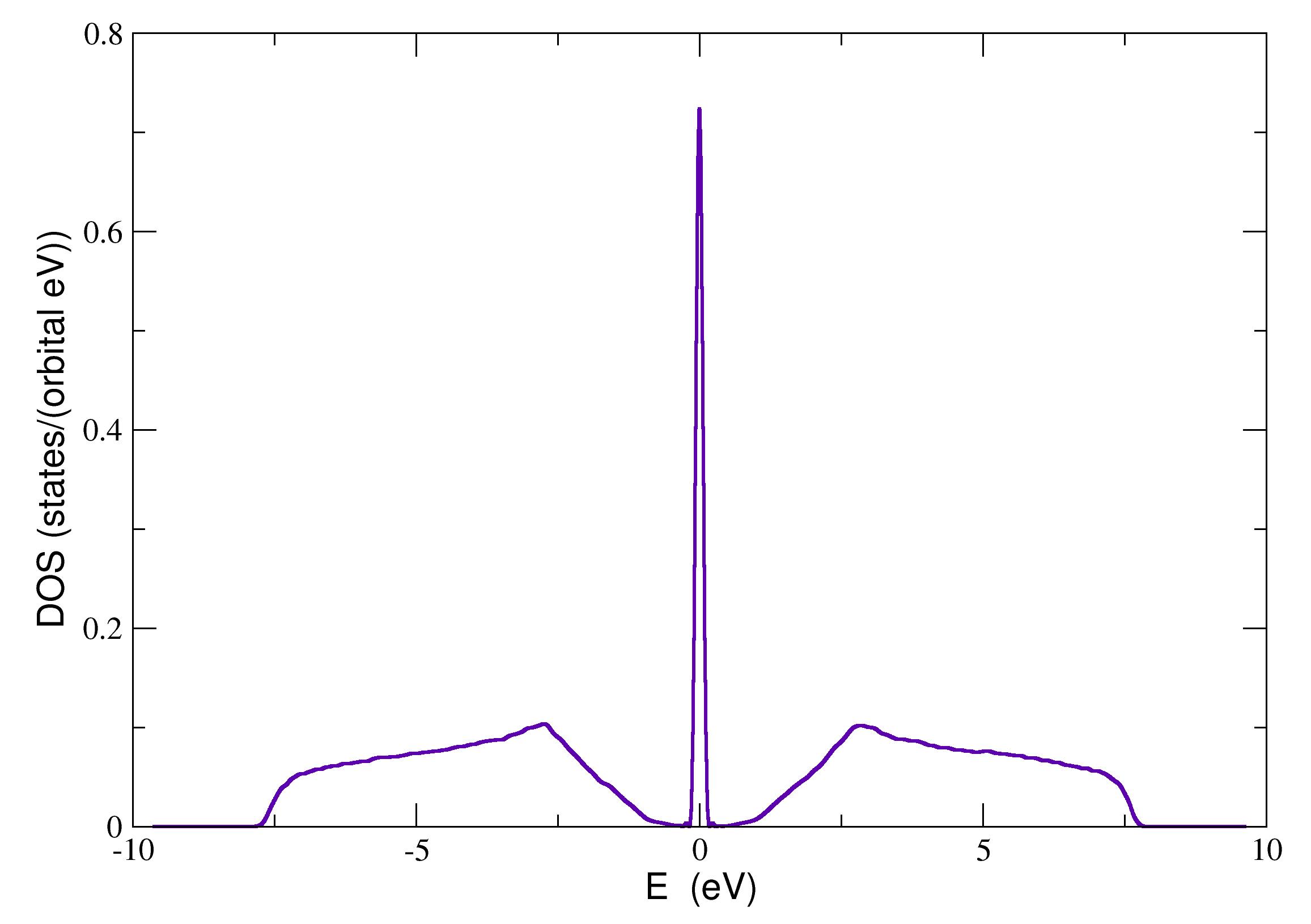
6. Disorder
KITE's general-purpose and user-friendly disorder implementation is one of its main features. The inclusion of disorder in a given system follows a simple but effective recipe: users select a default disorder type or define their own disorder patterns in the Python interface to create a bespoke disorder landscape. This information is passed on to KITEx and used to automatically perform the required modifications of on-site energies and hopping terms across the whole lattice on-the-fly.
Info
Disorder patterns are local modifications of the Hamiltonian that can be constricted to one unit cell or can connect neighboring unit cells. The local patterns are randomly distributed over the whole lattice with a desired concentration. This framework is thus ideal to simulate systems with random impurities/defects of specified types.
KITE handles both standard uncorrelated disorder (e.g., random on-site energies) and realistic short-range disorder (e.g., vacancies or impurity scattering centers distributed randomly over the lattice sites with a specified concentration). KITE can even handle multiple disorder patterns simultaneously.
Disorder implementation¶
After defining a regular lattice, disorder can be added to the system. KITE allows the user to select between on-site and structural disorder by choosing between predefined classes in the Python interface. The interface provides two different classes of disorder:
kite.Disorder- onsite disorder with two possible statistical distributionskite.StructuralDisorder- multi-orbital impurities and defects (including bond disorder) with a given concentration
On-site disorder¶
kite.Disorder adds randomly generated on-site terms at the sites of a desired sublattice drawn from one of the two statistical distributions:
- Gaussian
- Uniform
One can select a sublattice hosting a particular disorder, and specify the mean
value and the standard deviation of the selected distribution.
To include on-site disorder, one builds the pb.lattice and
use the following procedure:
# define an object based on the lattice
disorder = kite.Disorder(lattice)
# add Gaussian distributed disorder at all sites of a selected sublattice (A in this example)
disorder.add_disorder('A', 'Gaussian', mean, std)
In a single object it is possible to select multiple sublattices, each with its own disorder distribution
following the rule kite.Disorder.add_disorder('sublattice', 'type', mean, std). For example,
The final step consists of adding the disorder as an additional parameter into the
kite.config_system function:
A complete example that calculates the average density of states of graphene with different on-site disorder distributions on each sublattice is given below:
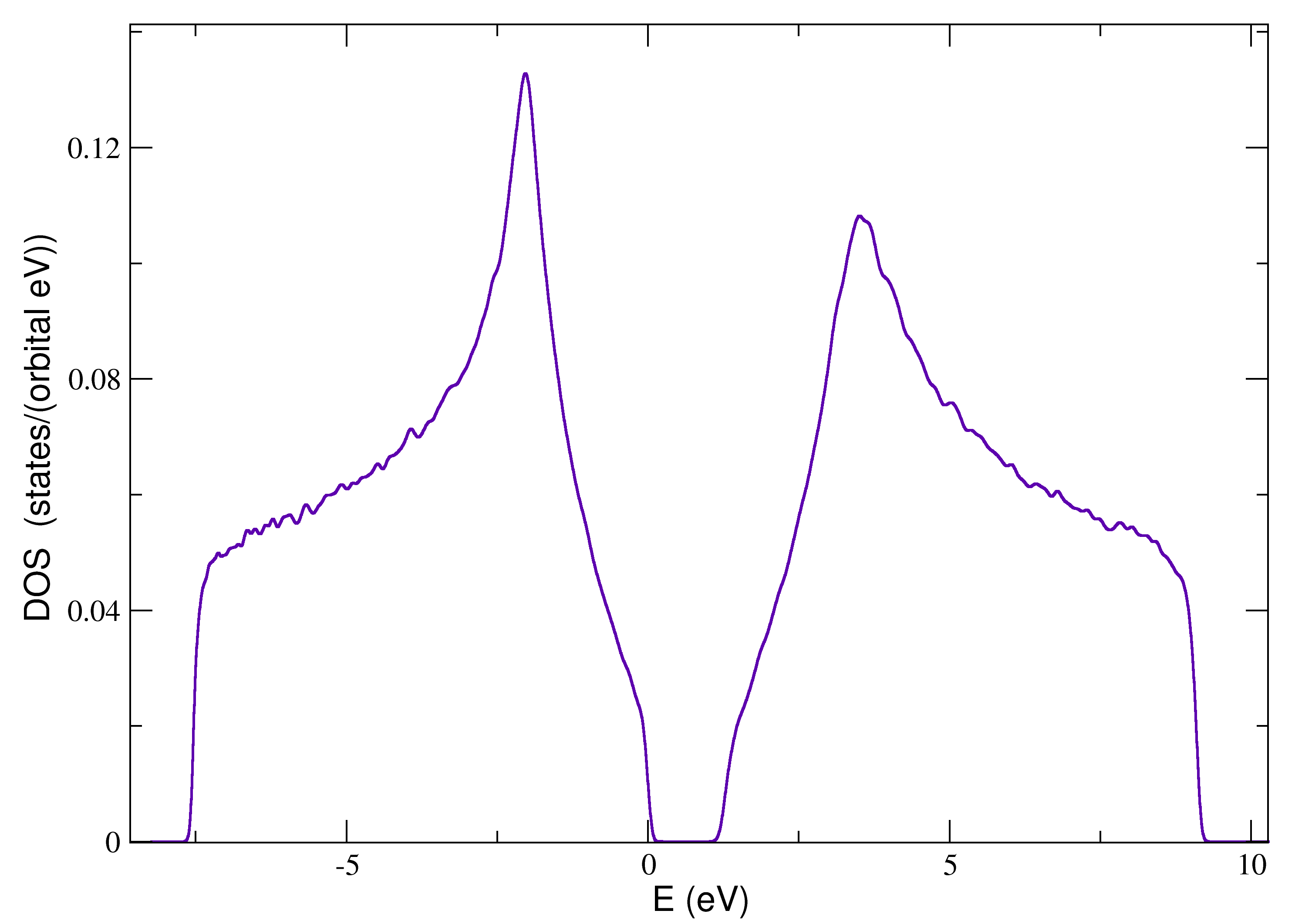
Structural disorder¶
The kite.StructuralDisorder class adds the possibility of selecting between two different short-range disorder types:
vacancy defects randomly distributed with a certain concentration over lattice sites on a selected sublattice,
and a more generic multi-orbital disorder which may combine on-site and hopping terms (also randomly distributed with a certain concentration).
Example 1: vacancy defects¶
To add vacancy defects with a given
concentration on a single sublattice, one uses the simple instruction:
struc_disorder = kite.StructuralDisorder(lattice, concentration=0.2)
struc_disorder.add_vacancy('B') # add a vacancy to a selected sublattice
Note
To distribute the vacancies on both sublattices (compensated or otherwise), one needs to treat each sublattice as a separate
object of the class kite.StructuralDisorder
Example 2: Mixed on-site/bond disorder¶
The following example illustrates KITE's most general type of short-range disorder, which includes both atomic defects (vacancies)
and bond modifications.
This type of disorder can be added as an object of the class kite.StructuralDisorder.
The procedure is analogous to adding a hopping term to the
Pybinding lattice object.
An important remark is in order. KITE's structural disorder functionality is rather general and allows users to create bespoke disorder landscapes for their simulations,
while benefiting from KITE's efficient domain decomposition algorithm. However, as mentioned in Sec. 2 - Lattice, it is assumed by default that all hopping terms in the model
connect sites that belong to either the same unit cell or neighboring cells that are separated by at most 2 lattice spacings along a given direction. This applies equally to the disorder patterns
as they are an inherent part of the lattice model. To relax this constraint, the users must adjust
the NGHOSTS parameter in kite/Src/Generic.hpp and recompile KITEx, otherwise an error message is output and the KITE program exits.
Let us begin by defining the sublattices that will constitute the disorder. In this case we are not restricted to a single unit cell:
# define a node in a unit cell [i, j] selecting a single sublattice
node0 = [[+0, +0], 'A']
node1 = [[+0, +0], 'B']
node2 = [[+1, +0], 'A']
node3 = [[+0, +1], 'B']
node4 = [[+0, +1], 'A']
node5 = [[-1, +1], 'B']
After defining a parent kite.StructuralDisorder object, we can construct the desired pattern:
# define an object based on the lattice with a certain concentration
struc_disorder = kite.StructuralDisorder(lattice, concentration=0.2)
struc_disorder.add_structural_disorder(
# add bond disorder in the form
# [from unit cell], 'sublattice_from', [to_unit_cell], 'sublattice_to', value:
(*node0, *node1, 0.5),
(*node1, *node2, 0.1),
(*node2, *node3, 0.5),
(*node3, *node4, 0.3),
(*node4, *node5, 0.4),
(*node5, *node0, 0.8),
# in this way we can add onsite disorder in the form [unit cell], 'sublattice', value
([+0, +0], 'B', 0.1)
)
# It is possible to add multiple different disorder types which
# should be forwarded to the config_system function as a list.
another_struc_disorder = kite.StructuralDisorder(lattice, concentration=0.6)
another_struc_disorder.add_structural_disorder(
(*node0, *node1, 0.05),
(*node4, *node5, 0.4),
(*node5, *node0, 0.02),
([+0, +0], 'A', 0.3)
)
Before exporting the settings to the HDF5-file, it is possible to define multiple disorder realizations which will be superimposed to the clean system.
The following script contains a brief example of how to configure the structural disorder:
with the resulting density of states: