2. Lattice
Note
A python script that generates KITE's input file requires few python packages that can be included with the following aliases
We will first construct a pb.Lattice using Pybinding
and calculate its density of states (DOS) using pb.Solver from Pybinding.
Info
If you are familiar with Pybinding, you can go directly to the next tutorial page.
Making a pb.Lattice¶
The pb.Lattice class from Pybinding carries the information about the TB model.
This includes
Pybinding also provides additional functionalities based on this real-space information. It can provide, for example, the reciprocal vectors and the Brillouin zone.
Defining the unit cell¶
As a simple example, let us construct a square lattice with a single lattice site. The following syntax can be used to define the primitive lattice vectors:
Adding lattice sites¶
We than add the desired lattice sites inside the unit cell (the same syntax can be used to add different orbitals in a given position or more sites in different sublattices):
Adding hoppings¶
By default, the main unit cell has the index [n1,n2] = [0, 0].
The hoppings between neighboring sites can be added with the simple syntax:
Here, the relative indices n1,n2 represent the number of integer steps - in units of the primitive lattice vectors - needed to reach a neighboring cell starting from the origin.
If the lattice has more than one sublattice, the hoppings can connect sites in the same unit cell.
Note
When adding the hopping (n, m) between sites n and m,
the conjugate hopping term (m, n) is added automatically. Pybinding does not allow the user to add them by hand.
Visualization¶
Now we can plot the pb.lattice and visualize the Brillouin zone:
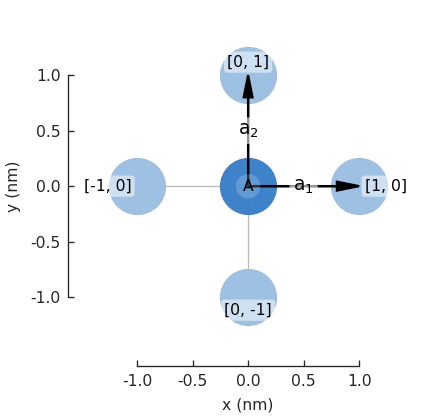
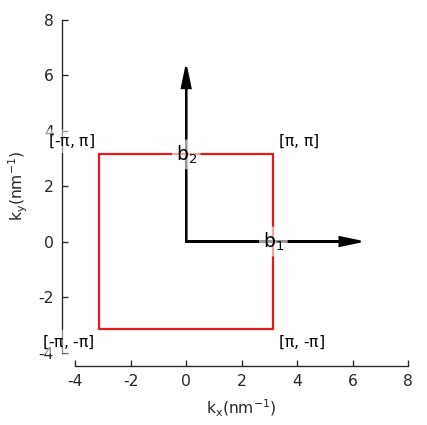
Examples
For a crystal with two atoms per unit cell, look in the Examples section. For other examples and pre-defined lattices consult the Pybinding documentation.
Using Pybinding's solver¶
Pybinding has build-in solvers for
- LAPACK (exact diagonalization) and
- ARPACK (targeted diagonalization of sparse matrices). To use any of these solvers, we need to first construct a model.
Building a pb.Model¶
The pb.Model class contains all the information of the structure we want to use in our calculation.
This structure can be larger than the unit cell (*stored in the pb.Lattice-class). It can also have specific geometries and other possible modifications of the original lattice.
Here, we will just double the unit cell in both directions in the pb.Model and add periodic boundary conditions:
pb.Model with
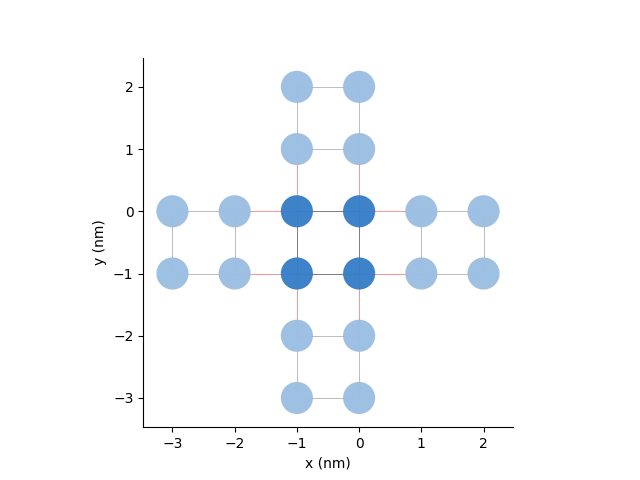
Defining a pb.Solver¶
The pb.Solver class takes a pb.Model class as input and prepares the system to perform a numerical calculation. We will use the LAPACK solver:
Band structure calculation¶
As an example, the band structure is calculated using the pb.Solver defined above.
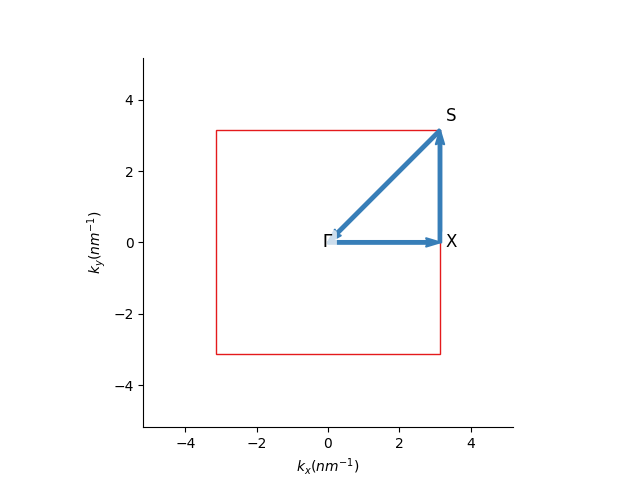
First, for a two-dimensional plot, we must define a path in the reciprocal space that connects the high symmetry points. Using the pb.Lattice build-in
method, the high symmetry points for the corners of a path can be found easily:
Then, we just pass these corners to the pb.Solver and visualize the result
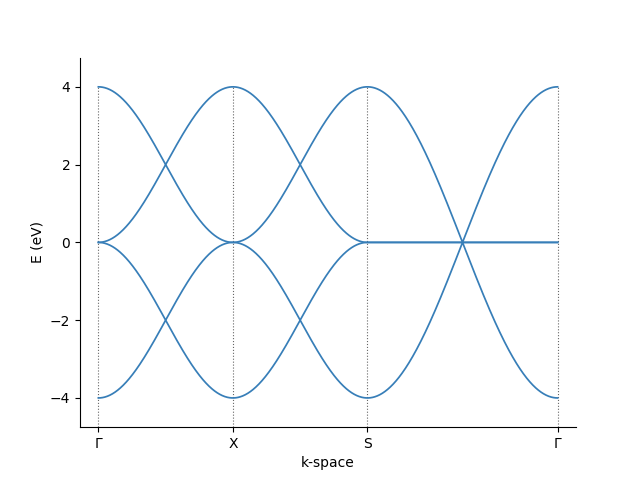
For more info about Pybinding's capabilities, look at its tutorial or API guide.
Summary of the code from this section